| Record Information |
|---|
| Version | 5.0 |
|---|
| Status | Detected and Quantified |
|---|
| Creation Date | 2005-11-16 15:48:42 UTC |
|---|
| Update Date | 2023-02-21 17:15:21 UTC |
|---|
| HMDB ID | HMDB0000965 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Hypotaurine |
|---|
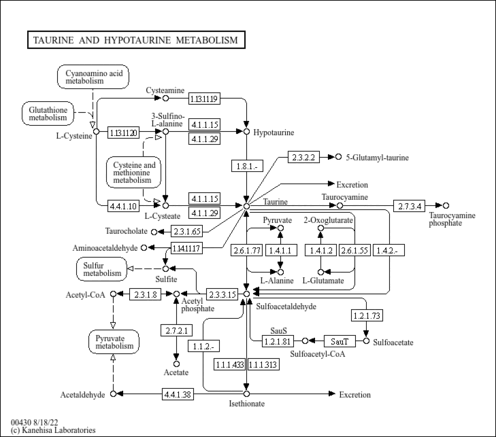
| Description | Hypotaurine belongs to the class of organic compounds known as sulfinic acids. Sulfinic acids are compounds containing a sulfinic acid functional group, with the general structure RS(=O)OH (R = organyl, not H). Hypotaurine exists in all living species, ranging from bacteria to humans. Within humans, hypotaurine participates in a number of enzymatic reactions. In particular, hypotaurine can be biosynthesized from cysteamine; which is catalyzed by the enzyme 2-aminoethanethiol dioxygenase. In addition, hypotaurine can be biosynthesized from 3-sulfinoalanine through its interaction with the enzyme cysteine sulfinic acid decarboxylase. In humans, hypotaurine is involved in taurine and hypotaurine metabolism. |
|---|
| Structure | InChI=1S/C2H7NO2S/c3-1-2-6(4)5/h1-3H2,(H,4,5) |
|---|
| Synonyms | | Value | Source |
|---|
| 2-Aminoethanesulfinic acid | ChEBI | | 2-Aminoethanesulfinate | Generator | | 2-Aminoethanesulphinate | Generator | | 2-Aminoethanesulphinic acid | Generator | | 2-Amino-ethanesulfinate | HMDB | | 2-Amino-ethanesulfinic acid | HMDB | | 2-Aminoethylsulfinate | HMDB | | 2-Aminoethylsulfinic acid | HMDB | | Cystaminesulfinate | HMDB | | Cystaminesulfinic acid | HMDB |
|
|---|
| Chemical Formula | C2H7NO2S |
|---|
| Average Molecular Weight | 109.147 |
|---|
| Monoisotopic Molecular Weight | 109.019749163 |
|---|
| IUPAC Name | 2-aminoethane-1-sulfinic acid |
|---|
| Traditional Name | hypotaurine |
|---|
| CAS Registry Number | 300-84-5 |
|---|
| SMILES | NCCS(O)=O |
|---|
| InChI Identifier | InChI=1S/C2H7NO2S/c3-1-2-6(4)5/h1-3H2,(H,4,5) |
|---|
| InChI Key | VVIUBCNYACGLLV-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as sulfinic acids. Sulfinic acids are compounds containing a sulfinic acid functional group, with the general structure RS(=O)OH (R = organyl, not H). |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Sulfinic acids and derivatives |
|---|
| Sub Class | Sulfinic acids |
|---|
| Direct Parent | Sulfinic acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Sulfinic acid
- Alkanesulfinic acid
- Alkanesulfinic acid or derivatives
- Organic nitrogen compound
- Organic oxygen compound
- Organopnictogen compound
- Organic oxide
- Hydrocarbon derivative
- Primary amine
- Organosulfur compound
- Organonitrogen compound
- Primary aliphatic amine
- Amine
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Not Available | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Molecular Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Experimental Chromatographic Properties | Experimental Collision Cross Sections |
|---|
| Predicted Molecular Properties | |
|---|
| Predicted Chromatographic Properties | Predicted Collision Cross SectionsPredicted Retention Times Underivatized| Chromatographic Method | Retention Time | Reference |
|---|
| Measured using a Waters Acquity ultraperformance liquid chromatography (UPLC) ethylene-bridged hybrid (BEH) C18 column (100 mm × 2.1 mm; 1.7 μmparticle diameter). Predicted by Afia on May 17, 2022. Predicted by Afia on May 17, 2022. | 1.2 minutes | 32390414 | | Predicted by Siyang on May 30, 2022 | 8.4843 minutes | 33406817 | | Predicted by Siyang using ReTip algorithm on June 8, 2022 | 6.73 minutes | 32390414 | | AjsUoB = Accucore 150 Amide HILIC with 10mM Ammonium Formate, 0.1% Formic Acid | 358.8 seconds | 40023050 | | Fem_Long = Waters ACQUITY UPLC HSS T3 C18 with Water:MeOH and 0.1% Formic Acid | 453.0 seconds | 40023050 | | Fem_Lipids = Ascentis Express C18 with (60:40 water:ACN):(90:10 IPA:ACN) and 10mM NH4COOH + 0.1% Formic Acid | 326.6 seconds | 40023050 | | Life_Old = Waters ACQUITY UPLC BEH C18 with Water:(20:80 acetone:ACN) and 0.1% Formic Acid | 56.2 seconds | 40023050 | | Life_New = RP Waters ACQUITY UPLC HSS T3 C18 with Water:(30:70 MeOH:ACN) and 0.1% Formic Acid | 226.4 seconds | 40023050 | | RIKEN = Waters ACQUITY UPLC BEH C18 with Water:ACN and 0.1% Formic Acid | 94.6 seconds | 40023050 | | Eawag_XBridgeC18 = XBridge C18 3.5u 2.1x50 mm with Water:MeOH and 0.1% Formic Acid | 256.1 seconds | 40023050 | | BfG_NTS_RP1 =Agilent Zorbax Eclipse Plus C18 (2.1 mm x 150 mm, 3.5 um) with Water:ACN and 0.1% Formic Acid | 222.6 seconds | 40023050 | | HILIC_BDD_2 = Merck SeQuant ZIC-HILIC with ACN(0.1% formic acid):water(16 mM ammonium formate) | 844.1 seconds | 40023050 | | UniToyama_Atlantis = RP Waters Atlantis T3 (2.1 x 150 mm, 5 um) with ACN:Water and 0.1% Formic Acid | 550.8 seconds | 40023050 | | BDD_C18 = Hypersil Gold 1.9µm C18 with Water:ACN and 0.1% Formic Acid | 35.5 seconds | 40023050 | | UFZ_Phenomenex = Kinetex Core-Shell C18 2.6 um, 3.0 x 100 mm, Phenomenex with Water:MeOH and 0.1% Formic Acid | 610.8 seconds | 40023050 | | SNU_RIKEN_POS = Waters ACQUITY UPLC BEH C18 with Water:ACN and 0.1% Formic Acid | 212.1 seconds | 40023050 | | RPMMFDA = Waters ACQUITY UPLC BEH C18 with Water:ACN and 0.1% Formic Acid | 306.1 seconds | 40023050 | | MTBLS87 = Merck SeQuant ZIC-pHILIC column with ACN:Water and :ammonium carbonate | 718.6 seconds | 40023050 | | KI_GIAR_zic_HILIC_pH2_7 = Merck SeQuant ZIC-HILIC with ACN:Water and 0.1% FA | 505.4 seconds | 40023050 | | Meister zic-pHILIC pH9.3 = Merck SeQuant ZIC-pHILIC column with ACN:Water 5mM NH4Ac pH9.3 and 5mM ammonium acetate in water | 349.4 seconds | 40023050 |
Predicted Kovats Retention IndicesUnderivatizedDerivatized| Derivative Name / Structure | SMILES | Kovats RI Value | Column Type | Reference |
|---|
| Hypotaurine,1TMS,isomer #1 | C[Si](C)(C)OS(=O)CCN | 1209.8 | Semi standard non polar | 33892256 | | Hypotaurine,1TMS,isomer #1 | C[Si](C)(C)OS(=O)CCN | 1330.2 | Standard non polar | 33892256 | | Hypotaurine,1TMS,isomer #1 | C[Si](C)(C)OS(=O)CCN | 1878.9 | Standard polar | 33892256 | | Hypotaurine,1TMS,isomer #2 | C[Si](C)(C)NCCS(=O)O | 1322.5 | Semi standard non polar | 33892256 | | Hypotaurine,1TMS,isomer #2 | C[Si](C)(C)NCCS(=O)O | 1393.6 | Standard non polar | 33892256 | | Hypotaurine,1TMS,isomer #2 | C[Si](C)(C)NCCS(=O)O | 1977.0 | Standard polar | 33892256 | | Hypotaurine,2TMS,isomer #1 | C[Si](C)(C)NCCS(=O)O[Si](C)(C)C | 1359.8 | Semi standard non polar | 33892256 | | Hypotaurine,2TMS,isomer #1 | C[Si](C)(C)NCCS(=O)O[Si](C)(C)C | 1590.1 | Standard non polar | 33892256 | | Hypotaurine,2TMS,isomer #1 | C[Si](C)(C)NCCS(=O)O[Si](C)(C)C | 1495.3 | Standard polar | 33892256 | | Hypotaurine,2TMS,isomer #2 | C[Si](C)(C)N(CCS(=O)O)[Si](C)(C)C | 1515.5 | Semi standard non polar | 33892256 | | Hypotaurine,2TMS,isomer #2 | C[Si](C)(C)N(CCS(=O)O)[Si](C)(C)C | 1688.9 | Standard non polar | 33892256 | | Hypotaurine,2TMS,isomer #2 | C[Si](C)(C)N(CCS(=O)O)[Si](C)(C)C | 1812.3 | Standard polar | 33892256 | | Hypotaurine,3TMS,isomer #1 | C[Si](C)(C)OS(=O)CCN([Si](C)(C)C)[Si](C)(C)C | 1558.5 | Semi standard non polar | 33892256 | | Hypotaurine,3TMS,isomer #1 | C[Si](C)(C)OS(=O)CCN([Si](C)(C)C)[Si](C)(C)C | 1809.8 | Standard non polar | 33892256 | | Hypotaurine,3TMS,isomer #1 | C[Si](C)(C)OS(=O)CCN([Si](C)(C)C)[Si](C)(C)C | 1513.7 | Standard polar | 33892256 | | Hypotaurine,1TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)OS(=O)CCN | 1425.6 | Semi standard non polar | 33892256 | | Hypotaurine,1TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)OS(=O)CCN | 1628.1 | Standard non polar | 33892256 | | Hypotaurine,1TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)OS(=O)CCN | 1958.6 | Standard polar | 33892256 | | Hypotaurine,1TBDMS,isomer #2 | CC(C)(C)[Si](C)(C)NCCS(=O)O | 1563.3 | Semi standard non polar | 33892256 | | Hypotaurine,1TBDMS,isomer #2 | CC(C)(C)[Si](C)(C)NCCS(=O)O | 1698.3 | Standard non polar | 33892256 | | Hypotaurine,1TBDMS,isomer #2 | CC(C)(C)[Si](C)(C)NCCS(=O)O | 2030.7 | Standard polar | 33892256 | | Hypotaurine,2TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)NCCS(=O)O[Si](C)(C)C(C)(C)C | 1791.1 | Semi standard non polar | 33892256 | | Hypotaurine,2TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)NCCS(=O)O[Si](C)(C)C(C)(C)C | 2135.9 | Standard non polar | 33892256 | | Hypotaurine,2TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)NCCS(=O)O[Si](C)(C)C(C)(C)C | 1738.4 | Standard polar | 33892256 | | Hypotaurine,2TBDMS,isomer #2 | CC(C)(C)[Si](C)(C)N(CCS(=O)O)[Si](C)(C)C(C)(C)C | 1945.0 | Semi standard non polar | 33892256 | | Hypotaurine,2TBDMS,isomer #2 | CC(C)(C)[Si](C)(C)N(CCS(=O)O)[Si](C)(C)C(C)(C)C | 2172.9 | Standard non polar | 33892256 | | Hypotaurine,2TBDMS,isomer #2 | CC(C)(C)[Si](C)(C)N(CCS(=O)O)[Si](C)(C)C(C)(C)C | 1915.8 | Standard polar | 33892256 | | Hypotaurine,3TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)OS(=O)CCN([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C | 2202.8 | Semi standard non polar | 33892256 | | Hypotaurine,3TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)OS(=O)CCN([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C | 2524.2 | Standard non polar | 33892256 | | Hypotaurine,3TBDMS,isomer #1 | CC(C)(C)[Si](C)(C)OS(=O)CCN([Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C | 1873.8 | Standard polar | 33892256 |
|
|---|
| General References | - Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, Nyati MK, Ahsan A, Kalyana-Sundaram S, Han B, Cao X, Byun J, Omenn GS, Ghosh D, Pennathur S, Alexander DC, Berger A, Shuster JR, Wei JT, Varambally S, Beecher C, Chinnaiyan AM: Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009 Feb 12;457(7231):910-4. doi: 10.1038/nature07762. [PubMed:19212411 ]
- Dominy J, Eller S, Dawson R Jr: Building biosynthetic schools: reviewing compartmentation of CNS taurine synthesis. Neurochem Res. 2004 Jan;29(1):97-103. [PubMed:14992267 ]
- Fontana M, Pecci L, Dupre S, Cavallini D: Antioxidant properties of sulfinates: protective effect of hypotaurine on peroxynitrite-dependent damage. Neurochem Res. 2004 Jan;29(1):111-6. [PubMed:14992269 ]
- Pitari G, Dupre S, Spirito A, Antonini G, Amicarelli F: Hypotaurine protection on cell damage by singlet oxygen. Adv Exp Med Biol. 2000;483:157-62. [PubMed:11787593 ]
- Grafe F, Wohlrab W, Neubert RH, Brandsch M: Functional characterization of sodium- and chloride-dependent taurine transport in human keratinocytes. Eur J Pharm Biopharm. 2004 Mar;57(2):337-41. [PubMed:15018993 ]
- Krieg RC, Uihlein D, Murthum T, Endlicher E, Hausmann F, Messmann H, Knuechel R: Improving the use of 5-aminolevulinic acid (ALA)-induced protoporphyrin IX (PPIX) for the gastrointestinal tract by esterification--an in vitro study. Cell Mol Biol (Noisy-le-grand). 2002 Dec;48(8):917-23. [PubMed:12699251 ]
- Guerin P, Menezo Y: Hypotaurine and taurine in gamete and embryo environments: de novo synthesis via the cysteine sulfinic acid pathway in oviduct cells. Zygote. 1995 Nov;3(4):333-43. [PubMed:8730898 ]
- Masuoka N, Yao K, Kinuta M, Ohta J, Wakimoto M, Ubuka T: High-performance liquid chromatographic determination of taurine and hypotaurine using 3,5-dinitrobenzoyl chloride as derivatizing reagent. J Chromatogr B Biomed Appl. 1994 Oct 3;660(1):31-5. [PubMed:7858721 ]
- Guerin P, Guillaud J, Menezo Y: Hypotaurine in spermatozoa and genital secretions and its production by oviduct epithelial cells in vitro. Hum Reprod. 1995 Apr;10(4):866-72. [PubMed:7650134 ]
- Holmes RP, Goodman HO, Shihabi ZK, Jarow JP: The taurine and hypotaurine content of human semen. J Androl. 1992 May-Jun;13(3):289-92. [PubMed:1601750 ]
- Mahadevan MM, Trounson AO: Removal of the cumulus oophorus from the human oocyte for in vitro fertilization. Fertil Steril. 1985 Feb;43(2):263-7. [PubMed:3967784 ]
- Elshenawy S, Pinney SE, Stuart T, Doulias PT, Zura G, Parry S, Elovitz MA, Bennett MJ, Bansal A, Strauss JF 3rd, Ischiropoulos H, Simmons RA: The Metabolomic Signature of the Placenta in Spontaneous Preterm Birth. Int J Mol Sci. 2020 Feb 4;21(3). pii: ijms21031043. doi: 10.3390/ijms21031043. [PubMed:32033212 ]
|
|---|