| Record Information |
|---|
| Version | 5.0 |
|---|
| Status | Detected and Quantified |
|---|
| Creation Date | 2005-11-16 15:48:42 UTC |
|---|
| Update Date | 2022-03-07 02:49:05 UTC |
|---|
| HMDB ID | HMDB0000722 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Lithocholyltaurine |
|---|
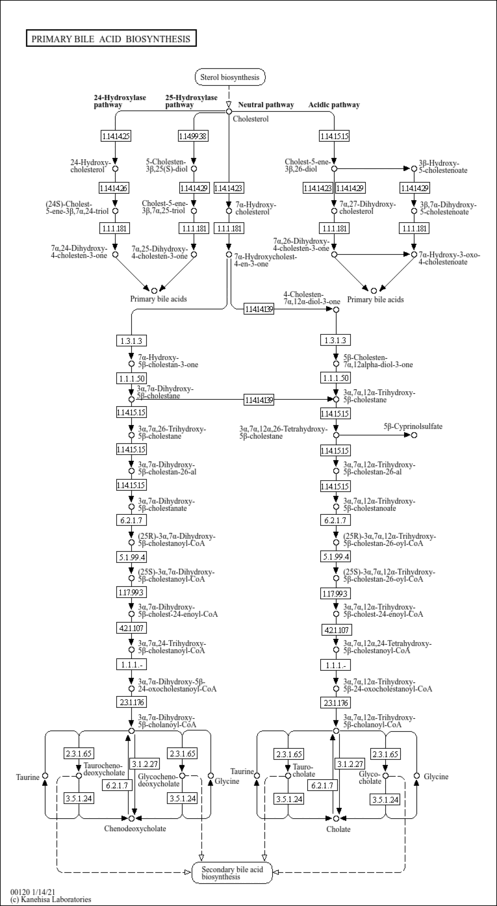
| Description | Lithocholyltaurine is a bile salt formed in the liver from lithocholic acid conjugation with taurine, usually as the sodium salt. It solubilizes fats for absorption and is itself absorbed. Lithocholic acid, a hydrophobic secondary bile acid, is well known to cause intrahepatic cholestasis. There have been extensive studies on the mechanisms of lithocholate-induced cholestasis in animals. Lithocholate diminishes both the bile acid-dependent and independent bile flow. In humans, elevated levels of lithocholic acid are found in patients with chronic cholestatic liver disease. Lithocholyltaurine impairs both the bile canalicular contractions and the canalicular bile secretion, possibly by acting directly on the canalicular membranes in lithocholyltaurine-induced cholestasis. Lithocholyltaurine induce acute cholestasis-associated with retrieval of the bile salt export pump. The bile salt export pump (BSEP) of hepatocyte secretes conjugated bile salts across the canalicular membrane in an ATP-dependent manner. Hepatic retention of bile acids may lead to liver injury by hepatocyte apoptosis and eventually deterioration of cholestatic liver diseases. One mechanism of induced apoptosis by lithocholyltaurine is the induction of transcriptional activity of AP-1 (activation protein-1). (PMID: 16981261 , 15763547 , 16332456 , 18164257 ). |
|---|
| Structure | [H][C@@]1(CC[C@@]2([H])[C@]3([H])CC[C@]4([H])C[C@H](O)CC[C@]4(C)[C@@]3([H])CC[C@]12C)[C@H](C)CCC(=O)NCCS(O)(=O)=O InChI=1S/C26H45NO5S/c1-17(4-9-24(29)27-14-15-33(30,31)32)21-7-8-22-20-6-5-18-16-19(28)10-12-25(18,2)23(20)11-13-26(21,22)3/h17-23,28H,4-16H2,1-3H3,(H,27,29)(H,30,31,32)/t17-,18-,19-,20+,21-,22+,23+,25+,26-/m1/s1 |
|---|
| Synonyms | | Value | Source |
|---|
| Taurolithocholate | ChEBI | | Taurolithocholic acid | Kegg | | Taurine lithocholate | HMDB | | Acid, taurolithocholic | HMDB | | Taurolithocholic acid, monosodium salt | HMDB | | Lithocholate, taurine | HMDB | | 3alpha-Hydroxy-5beta-cholanoyltaurine | HMDB | | 3alpha-Hydroxy-N-(2-sulfoethyl)-5beta-cholan-24-amide | HMDB | | 3Α-hydroxy-5β-cholanoyltaurine | HMDB | | 3Α-hydroxy-N-(2-sulfoethyl)-5β-cholan-24-amide | HMDB | | Lithocholic acid taurine conjugate | HMDB | | Lithocholyltaurine | ChEBI |
|
|---|
| Chemical Formula | C26H45NO5S |
|---|
| Average Molecular Weight | 483.71 |
|---|
| Monoisotopic Molecular Weight | 483.301844727 |
|---|
| IUPAC Name | 2-[(4R)-4-[(1S,2S,5R,7R,10R,11S,14R,15R)-5-hydroxy-2,15-dimethyltetracyclo[8.7.0.0^{2,7}.0^{11,15}]heptadecan-14-yl]pentanamido]ethane-1-sulfonic acid |
|---|
| Traditional Name | 2-[(4R)-4-[(1S,2S,5R,7R,10R,11S,14R,15R)-5-hydroxy-2,15-dimethyltetracyclo[8.7.0.0^{2,7}.0^{11,15}]heptadecan-14-yl]pentanamido]ethanesulfonic acid |
|---|
| CAS Registry Number | 516-90-5 |
|---|
| SMILES | [H][C@@]1(CC[C@@]2([H])[C@]3([H])CC[C@]4([H])C[C@H](O)CC[C@]4(C)[C@@]3([H])CC[C@]12C)[C@H](C)CCC(=O)NCCS(O)(=O)=O |
|---|
| InChI Identifier | InChI=1S/C26H45NO5S/c1-17(4-9-24(29)27-14-15-33(30,31)32)21-7-8-22-20-6-5-18-16-19(28)10-12-25(18,2)23(20)11-13-26(21,22)3/h17-23,28H,4-16H2,1-3H3,(H,27,29)(H,30,31,32)/t17-,18-,19-,20+,21-,22+,23+,25+,26-/m1/s1 |
|---|
| InChI Key | QBYUNVOYXHFVKC-GBURMNQMSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as taurinated bile acids and derivatives. These are bile acid derivatives containing a taurine conjugated to the bile acid moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Steroids and steroid derivatives |
|---|
| Sub Class | Bile acids, alcohols and derivatives |
|---|
| Direct Parent | Taurinated bile acids and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Taurinated bile acid
- Hydroxy bile acid, alcohol, or derivatives
- Monohydroxy bile acid, alcohol, or derivatives
- 3-hydroxysteroid
- Hydroxysteroid
- 3-alpha-hydroxysteroid
- Fatty amide
- N-acyl-amine
- Fatty acyl
- Sulfonyl
- Organosulfonic acid
- Organosulfonic acid or derivatives
- Organic sulfonic acid or derivatives
- Alkanesulfonic acid
- Cyclic alcohol
- Secondary carboxylic acid amide
- Secondary alcohol
- Carboxamide group
- Carboxylic acid derivative
- Organic oxide
- Organopnictogen compound
- Organic oxygen compound
- Organic nitrogen compound
- Organonitrogen compound
- Organosulfur compound
- Organooxygen compound
- Carbonyl group
- Hydrocarbon derivative
- Alcohol
- Aliphatic homopolycyclic compound
|
|---|
| Molecular Framework | Aliphatic homopolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Physiological effect | Not Available |
|---|
| Disposition | |
|---|
| Process | |
|---|
| Role | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Molecular Properties | | Property | Value | Reference |
|---|
| Melting Point | 212 - 213 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Experimental Chromatographic Properties | Not Available |
|---|
| Predicted Molecular Properties | |
|---|
| Predicted Chromatographic Properties | Predicted Collision Cross SectionsPredicted Retention Times Underivatized| Chromatographic Method | Retention Time | Reference |
|---|
| Measured using a Waters Acquity ultraperformance liquid chromatography (UPLC) ethylene-bridged hybrid (BEH) C18 column (100 mm × 2.1 mm; 1.7 μmparticle diameter). Predicted by Afia on May 17, 2022. Predicted by Afia on May 17, 2022. | 8.22 minutes | 32390414 | | Predicted by Siyang on May 30, 2022 | 15.9534 minutes | 33406817 | | Predicted by Siyang using ReTip algorithm on June 8, 2022 | 2.78 minutes | 32390414 | | AjsUoB = Accucore 150 Amide HILIC with 10mM Ammonium Formate, 0.1% Formic Acid | 79.6 seconds | 40023050 | | Fem_Long = Waters ACQUITY UPLC HSS T3 C18 with Water:MeOH and 0.1% Formic Acid | 3093.6 seconds | 40023050 | | Fem_Lipids = Ascentis Express C18 with (60:40 water:ACN):(90:10 IPA:ACN) and 10mM NH4COOH + 0.1% Formic Acid | 213.3 seconds | 40023050 | | Life_Old = Waters ACQUITY UPLC BEH C18 with Water:(20:80 acetone:ACN) and 0.1% Formic Acid | 231.7 seconds | 40023050 | | Life_New = RP Waters ACQUITY UPLC HSS T3 C18 with Water:(30:70 MeOH:ACN) and 0.1% Formic Acid | 173.0 seconds | 40023050 | | RIKEN = Waters ACQUITY UPLC BEH C18 with Water:ACN and 0.1% Formic Acid | 652.6 seconds | 40023050 | | Eawag_XBridgeC18 = XBridge C18 3.5u 2.1x50 mm with Water:MeOH and 0.1% Formic Acid | 638.0 seconds | 40023050 | | BfG_NTS_RP1 =Agilent Zorbax Eclipse Plus C18 (2.1 mm x 150 mm, 3.5 um) with Water:ACN and 0.1% Formic Acid | 656.5 seconds | 40023050 | | HILIC_BDD_2 = Merck SeQuant ZIC-HILIC with ACN(0.1% formic acid):water(16 mM ammonium formate) | 325.0 seconds | 40023050 | | UniToyama_Atlantis = RP Waters Atlantis T3 (2.1 x 150 mm, 5 um) with ACN:Water and 0.1% Formic Acid | 1245.2 seconds | 40023050 | | BDD_C18 = Hypersil Gold 1.9µm C18 with Water:ACN and 0.1% Formic Acid | 555.2 seconds | 40023050 | | UFZ_Phenomenex = Kinetex Core-Shell C18 2.6 um, 3.0 x 100 mm, Phenomenex with Water:MeOH and 0.1% Formic Acid | 1708.9 seconds | 40023050 | | SNU_RIKEN_POS = Waters ACQUITY UPLC BEH C18 with Water:ACN and 0.1% Formic Acid | 387.6 seconds | 40023050 | | RPMMFDA = Waters ACQUITY UPLC BEH C18 with Water:ACN and 0.1% Formic Acid | 450.4 seconds | 40023050 | | MTBLS87 = Merck SeQuant ZIC-pHILIC column with ACN:Water and :ammonium carbonate | 230.3 seconds | 40023050 | | KI_GIAR_zic_HILIC_pH2_7 = Merck SeQuant ZIC-HILIC with ACN:Water and 0.1% FA | 254.4 seconds | 40023050 | | Meister zic-pHILIC pH9.3 = Merck SeQuant ZIC-pHILIC column with ACN:Water 5mM NH4Ac pH9.3 and 5mM ammonium acetate in water | 22.0 seconds | 40023050 |
Predicted Kovats Retention IndicesUnderivatizedDerivatized| Derivative Name / Structure | SMILES | Kovats RI Value | Column Type | Reference |
|---|
| Lithocholyltaurine,1TMS,isomer #1 | C[C@H](CCC(=O)NCCS(=O)(=O)O)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O[Si](C)(C)C)CC[C@]4(C)[C@H]3CC[C@@]21C | 4184.8 | Semi standard non polar | 33892256 | | Lithocholyltaurine,1TMS,isomer #2 | C[C@H](CCC(=O)NCCS(=O)(=O)O[Si](C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O)CC[C@]4(C)[C@H]3CC[C@@]21C | 4227.4 | Semi standard non polar | 33892256 | | Lithocholyltaurine,1TMS,isomer #3 | C[C@H](CCC(=O)N(CCS(=O)(=O)O)[Si](C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O)CC[C@]4(C)[C@H]3CC[C@@]21C | 4181.5 | Semi standard non polar | 33892256 | | Lithocholyltaurine,2TMS,isomer #1 | C[C@H](CCC(=O)NCCS(=O)(=O)O[Si](C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O[Si](C)(C)C)CC[C@]4(C)[C@H]3CC[C@@]21C | 4245.0 | Semi standard non polar | 33892256 | | Lithocholyltaurine,2TMS,isomer #1 | C[C@H](CCC(=O)NCCS(=O)(=O)O[Si](C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O[Si](C)(C)C)CC[C@]4(C)[C@H]3CC[C@@]21C | 4178.9 | Standard non polar | 33892256 | | Lithocholyltaurine,2TMS,isomer #1 | C[C@H](CCC(=O)NCCS(=O)(=O)O[Si](C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O[Si](C)(C)C)CC[C@]4(C)[C@H]3CC[C@@]21C | 4915.9 | Standard polar | 33892256 | | Lithocholyltaurine,2TMS,isomer #2 | C[C@H](CCC(=O)N(CCS(=O)(=O)O)[Si](C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O[Si](C)(C)C)CC[C@]4(C)[C@H]3CC[C@@]21C | 4180.2 | Semi standard non polar | 33892256 | | Lithocholyltaurine,2TMS,isomer #2 | C[C@H](CCC(=O)N(CCS(=O)(=O)O)[Si](C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O[Si](C)(C)C)CC[C@]4(C)[C@H]3CC[C@@]21C | 4272.3 | Standard non polar | 33892256 | | Lithocholyltaurine,2TMS,isomer #2 | C[C@H](CCC(=O)N(CCS(=O)(=O)O)[Si](C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O[Si](C)(C)C)CC[C@]4(C)[C@H]3CC[C@@]21C | 4998.5 | Standard polar | 33892256 | | Lithocholyltaurine,2TMS,isomer #3 | C[C@H](CCC(=O)N(CCS(=O)(=O)O[Si](C)(C)C)[Si](C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O)CC[C@]4(C)[C@H]3CC[C@@]21C | 4237.9 | Semi standard non polar | 33892256 | | Lithocholyltaurine,2TMS,isomer #3 | C[C@H](CCC(=O)N(CCS(=O)(=O)O[Si](C)(C)C)[Si](C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O)CC[C@]4(C)[C@H]3CC[C@@]21C | 4251.6 | Standard non polar | 33892256 | | Lithocholyltaurine,2TMS,isomer #3 | C[C@H](CCC(=O)N(CCS(=O)(=O)O[Si](C)(C)C)[Si](C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O)CC[C@]4(C)[C@H]3CC[C@@]21C | 4866.2 | Standard polar | 33892256 | | Lithocholyltaurine,3TMS,isomer #1 | C[C@H](CCC(=O)N(CCS(=O)(=O)O[Si](C)(C)C)[Si](C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O[Si](C)(C)C)CC[C@]4(C)[C@H]3CC[C@@]21C | 4193.9 | Semi standard non polar | 33892256 | | Lithocholyltaurine,3TMS,isomer #1 | C[C@H](CCC(=O)N(CCS(=O)(=O)O[Si](C)(C)C)[Si](C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O[Si](C)(C)C)CC[C@]4(C)[C@H]3CC[C@@]21C | 4399.0 | Standard non polar | 33892256 | | Lithocholyltaurine,3TMS,isomer #1 | C[C@H](CCC(=O)N(CCS(=O)(=O)O[Si](C)(C)C)[Si](C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O[Si](C)(C)C)CC[C@]4(C)[C@H]3CC[C@@]21C | 4790.6 | Standard polar | 33892256 | | Lithocholyltaurine,1TBDMS,isomer #1 | C[C@H](CCC(=O)NCCS(=O)(=O)O)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O[Si](C)(C)C(C)(C)C)CC[C@]4(C)[C@H]3CC[C@@]21C | 4405.5 | Semi standard non polar | 33892256 | | Lithocholyltaurine,1TBDMS,isomer #2 | C[C@H](CCC(=O)NCCS(=O)(=O)O[Si](C)(C)C(C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O)CC[C@]4(C)[C@H]3CC[C@@]21C | 4411.4 | Semi standard non polar | 33892256 | | Lithocholyltaurine,1TBDMS,isomer #3 | C[C@H](CCC(=O)N(CCS(=O)(=O)O)[Si](C)(C)C(C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O)CC[C@]4(C)[C@H]3CC[C@@]21C | 4416.6 | Semi standard non polar | 33892256 | | Lithocholyltaurine,2TBDMS,isomer #1 | C[C@H](CCC(=O)NCCS(=O)(=O)O[Si](C)(C)C(C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O[Si](C)(C)C(C)(C)C)CC[C@]4(C)[C@H]3CC[C@@]21C | 4659.9 | Semi standard non polar | 33892256 | | Lithocholyltaurine,2TBDMS,isomer #1 | C[C@H](CCC(=O)NCCS(=O)(=O)O[Si](C)(C)C(C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O[Si](C)(C)C(C)(C)C)CC[C@]4(C)[C@H]3CC[C@@]21C | 4723.5 | Standard non polar | 33892256 | | Lithocholyltaurine,2TBDMS,isomer #1 | C[C@H](CCC(=O)NCCS(=O)(=O)O[Si](C)(C)C(C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O[Si](C)(C)C(C)(C)C)CC[C@]4(C)[C@H]3CC[C@@]21C | 4998.6 | Standard polar | 33892256 | | Lithocholyltaurine,2TBDMS,isomer #2 | C[C@H](CCC(=O)N(CCS(=O)(=O)O)[Si](C)(C)C(C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O[Si](C)(C)C(C)(C)C)CC[C@]4(C)[C@H]3CC[C@@]21C | 4617.9 | Semi standard non polar | 33892256 | | Lithocholyltaurine,2TBDMS,isomer #2 | C[C@H](CCC(=O)N(CCS(=O)(=O)O)[Si](C)(C)C(C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O[Si](C)(C)C(C)(C)C)CC[C@]4(C)[C@H]3CC[C@@]21C | 4767.1 | Standard non polar | 33892256 | | Lithocholyltaurine,2TBDMS,isomer #2 | C[C@H](CCC(=O)N(CCS(=O)(=O)O)[Si](C)(C)C(C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O[Si](C)(C)C(C)(C)C)CC[C@]4(C)[C@H]3CC[C@@]21C | 5094.0 | Standard polar | 33892256 | | Lithocholyltaurine,2TBDMS,isomer #3 | C[C@H](CCC(=O)N(CCS(=O)(=O)O[Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O)CC[C@]4(C)[C@H]3CC[C@@]21C | 4679.1 | Semi standard non polar | 33892256 | | Lithocholyltaurine,2TBDMS,isomer #3 | C[C@H](CCC(=O)N(CCS(=O)(=O)O[Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O)CC[C@]4(C)[C@H]3CC[C@@]21C | 4782.9 | Standard non polar | 33892256 | | Lithocholyltaurine,2TBDMS,isomer #3 | C[C@H](CCC(=O)N(CCS(=O)(=O)O[Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O)CC[C@]4(C)[C@H]3CC[C@@]21C | 4972.7 | Standard polar | 33892256 | | Lithocholyltaurine,3TBDMS,isomer #1 | C[C@H](CCC(=O)N(CCS(=O)(=O)O[Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O[Si](C)(C)C(C)(C)C)CC[C@]4(C)[C@H]3CC[C@@]21C | 4860.0 | Semi standard non polar | 33892256 | | Lithocholyltaurine,3TBDMS,isomer #1 | C[C@H](CCC(=O)N(CCS(=O)(=O)O[Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O[Si](C)(C)C(C)(C)C)CC[C@]4(C)[C@H]3CC[C@@]21C | 5176.9 | Standard non polar | 33892256 | | Lithocholyltaurine,3TBDMS,isomer #1 | C[C@H](CCC(=O)N(CCS(=O)(=O)O[Si](C)(C)C(C)(C)C)[Si](C)(C)C(C)(C)C)[C@H]1CC[C@H]2[C@@H]3CC[C@@H]4C[C@H](O[Si](C)(C)C(C)(C)C)CC[C@]4(C)[C@H]3CC[C@@]21C | 4897.3 | Standard polar | 33892256 |
|
|---|
| GC-MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - Lithocholyltaurine GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Lithocholyltaurine GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Lithocholyltaurine GC-MS (TMS_1_1) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Lithocholyltaurine GC-MS (TMS_1_2) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Lithocholyltaurine GC-MS (TMS_1_3) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Lithocholyltaurine GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Lithocholyltaurine GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - Lithocholyltaurine GC-MS (TBDMS_1_3) - 70eV, Positive | Not Available | 2021-11-05 | Wishart Lab | View Spectrum |
MS/MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Experimental LC-MS/MS | LC-MS/MS Spectrum - Lithocholyltaurine 20V, Negative-QTOF | splash10-001i-0000900000-62d017c1c686f7266fb0 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Lithocholyltaurine 20V, Positive-QTOF | splash10-014i-0000900000-370b7bde61cf15901a05 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Lithocholyltaurine 20V, Negative-QTOF | splash10-001i-0000900000-a35b80c6756ffe4a9735 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Lithocholyltaurine 40V, Negative-QTOF | splash10-001i-0000900000-7d2d59d9367fc3097f6a | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Lithocholyltaurine 10V, Negative-QTOF | splash10-001i-0000900000-cb0b19ca18aeb20850bf | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Lithocholyltaurine 40V, Negative-QTOF | splash10-001i-0000900000-d527b7971f2a10869476 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Lithocholyltaurine 10V, Negative-QTOF | splash10-001i-0000900000-a8f1d5fe4d9c4ae2b973 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Lithocholyltaurine 10V, Negative-QTOF | splash10-001i-0000900000-b7f7991f18a8750f971a | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Lithocholyltaurine 20V, Negative-QTOF | splash10-001i-0000900000-e08a1b85b998b206ce04 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Lithocholyltaurine 40V, Negative-QTOF | splash10-001i-0000900000-8f83f6d43ddf7e3ef518 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Lithocholyltaurine 10V, Positive-QTOF | splash10-014i-0000900000-3c99186da6a80b384824 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Lithocholyltaurine 20V, Positive-QTOF | splash10-014i-0536900000-3c8a96d3193eee3c45e7 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Lithocholyltaurine 10V, Positive-QTOF | splash10-014i-0000900000-d532dc016f756e101955 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Lithocholyltaurine 20V, Positive-QTOF | splash10-014i-0000900000-14b55a77ecc8c685282f | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Lithocholyltaurine 10V, Positive-QTOF | splash10-014i-0001900000-4faf258ccbd40728478e | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Lithocholyltaurine 40V, Positive-QTOF | splash10-00fu-3986000000-6221debd3d96ad748cf0 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Lithocholyltaurine 40V, Positive-QTOF | splash10-004i-2920000000-8d3c5fefb8cdf48324cc | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Lithocholyltaurine 10V, Positive-QTOF | splash10-014i-0000900000-8e6cbe0112d70a9c7554 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Lithocholyltaurine 20V, Positive-QTOF | splash10-016r-3946800000-c1a033a5e37851135b55 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Lithocholyltaurine 40V, Positive-QTOF | splash10-0571-5930000000-f06779a0e2a6b59baf2e | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Lithocholyltaurine 20V, Positive-QTOF | splash10-014i-0316900000-c834f65be00b11b05256 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Lithocholyltaurine 10V, Positive-QTOF | splash10-014i-0001900000-0dbbc8861a1b2d702ccc | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Lithocholyltaurine 20V, Positive-QTOF | splash10-014i-1647900000-1959f275d5a4ff01e360 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Lithocholyltaurine 40V, Positive-QTOF | splash10-004i-3920000000-79daf69ae12d0ea902f0 | 2021-09-20 | HMDB team, MONA | View Spectrum | | Experimental LC-MS/MS | LC-MS/MS Spectrum - Lithocholyltaurine 10V, Positive-QTOF | splash10-014i-0000900000-07e531c1684fe7ea6a4b | 2021-09-20 | HMDB team, MONA | View Spectrum |
NMR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Predicted 1D NMR | 13C NMR Spectrum (1D, 100 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 100 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 200 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 200 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 300 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 300 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 400 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 500 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 600 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 700 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 700 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 800 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 800 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 900 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 900 MHz, H2O, predicted) | 2022-08-22 | Wishart Lab | View Spectrum |
|
|---|
| General References | - Tadano T, Kanoh M, Matsumoto M, Sakamoto K, Kamano T: Studies of serum and feces bile acids determination by gas chromatography-mass spectrometry. Rinsho Byori. 2006 Feb;54(2):103-10. [PubMed:16548228 ]
- Lee BL, New AL, Ong CN: Comparative analysis of conjugated bile acids in human serum using high-performance liquid chromatography and capillary electrophoresis. J Chromatogr B Biomed Sci Appl. 1997 Dec 19;704(1-2):35-42. [PubMed:9518169 ]
- Hayashi H, Takada T, Suzuki H, Onuki R, Hofmann AF, Sugiyama Y: Transport by vesicles of glycine- and taurine-conjugated bile salts and taurolithocholate 3-sulfate: a comparison of human BSEP with rat Bsep. Biochim Biophys Acta. 2005 Dec 30;1738(1-3):54-62. Epub 2005 Nov 15. [PubMed:16332456 ]
- Watanabe N, Kagawa T, Kojima S, Takashimizu S, Nagata N, Nishizaki Y, Mine T: Taurolithocholate impairs bile canalicular motility and canalicular bile secretion in isolated rat hepatocyte couplets. World J Gastroenterol. 2006 Sep 7;12(33):5320-5. [PubMed:16981261 ]
- Crocenzi FA, Basiglio CL, Perez LM, Portesio MS, Pozzi EJ, Roma MG: Silibinin prevents cholestasis-associated retrieval of the bile salt export pump, Bsep, in isolated rat hepatocyte couplets: possible involvement of cAMP. Biochem Pharmacol. 2005 Apr 1;69(7):1113-20. [PubMed:15763547 ]
- Pusl T, Vennegeerts T, Wimmer R, Denk GU, Beuers U, Rust C: Tauroursodeoxycholic acid reduces bile acid-induced apoptosis by modulation of AP-1. Biochem Biophys Res Commun. 2008 Feb 29;367(1):208-12. doi: 10.1016/j.bbrc.2007.12.122. Epub 2007 Dec 27. [PubMed:18164257 ]
|
|---|